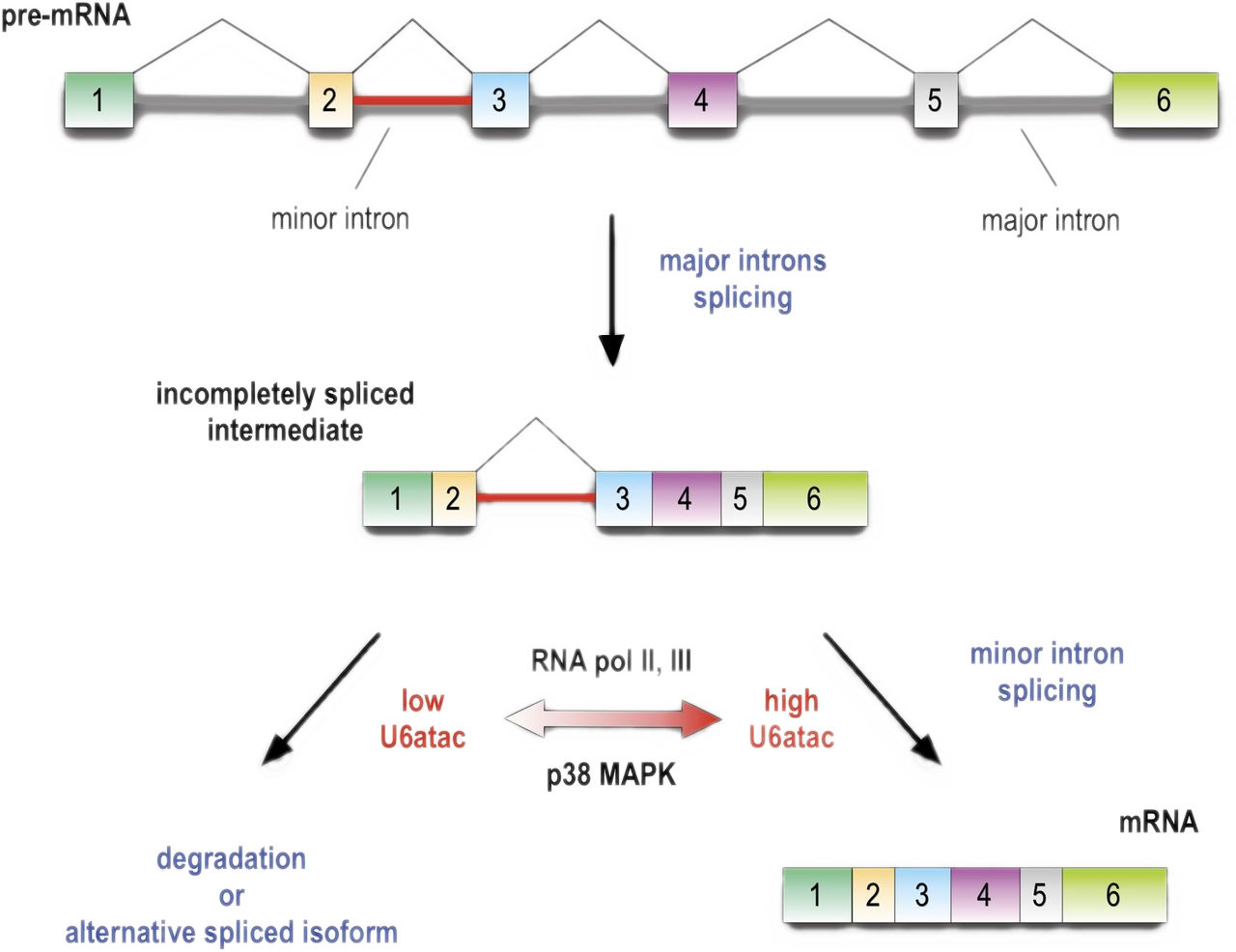
Minor introns are embedded molecular switches regulated by the abundance of highly unstable U6atac snRNA
In addition to containing typical (‘major’) introns, several hundred human genes also contain a single ‘minor’ intron, and a minor spliceosome is needed to remove it. Minor introns occur in many highly conserved genes, but they are often inefficiently spliced. This means that the resulting mRNA transcripts may not be translated into proteins—which is puzzling given that these proteins perform important roles within the cell.
In a new study, investigators found that a scarce, small RNA, called U6atac, controls the expression of hundreds of genes that have critical functions in cell growth, cell-cycle control, and global control of physiology (through regulation of several transcription regulators, ion channels, signaling proteins, and DNA damage-repair proteins).
The major spliceosome is by far the most abundant, and its minor counterpart (which has similar but not identical components) is often wholly disregarded.
This apparently superfluous secondary mechanism would appear to be a prime target for evolution to weed out — mRNAs produced from genes that have a minor intron are not ready until all their introns, both major and minor, are spliced. Thus a single inefficiently spliced minor intron can hold up expression for an entire gene.
When minor spliceosome activity is reduced, the minor introns are retained in the mRNA. This signals the mRNA for degradation, limiting the expression of genes that contain minor introns.
Serendipitously, inhibition of transcription was tested for an effect on snRNPs and soon showed a sharp drop in cellular U6atac — also the catalytic component of the minor spliceosome. The instability of the molecule helps it act as a molecular switch, since it’s already one of the rarest snRNAs in the cell. Surely stop transcribing it, and due to its scarcity the whole process of minor splicing will rapidly come to a standstill?
To verify, the researchers knocked down U6atac and then sequenced genome-wide RNA. More interestingly than a straightforward silencing, each minor intron responded differently, some being more sensitive to the loss of U6atac (and more inefficiently spliced), explaining the poor expression of mRNA from those genes.
Low U6atac levels within cells limit the rate of minor intron splicing, and thus the expression of important genes containing those minor introns.
Surveying various conditions that increase intracellular U6atac, it was found that cell stress, activating the p38MAPK pathway, causes a very large and rapid increase in U6atac and a huge accompanying enhancement in the splicing of those minor introns that otherwise do so very inefficiently.
Splicing of minor introns, typically one amidst several major introns, is dependent on the limiting level of U6atac snRNP in cells. Low abundance and fast turnover of U6atac results in incompletely spliced pre-mRNAs that are either degraded or switch into spliced isoforms that do not require the minor spliceosome. While transcription attenuation rapidly lowers U6atac level and limits minor intron-containing gene’s expression, activated p38MAPK rapidly stabilizes U6atac, increasing its level and enhancing minor splicing and production of full-length mRNAs.
Cell stress as transduced by p38MAPK can mean released inflammatory cytokines, UV radiation, heat, and osmotic shocks, giving the signalling molecule an important role in cellular growth and differentiation, apoptosis, cancer, and autophagy.
Together, these results imply that the minor spliceosome is used as a valve that can help cells to adapt to stress and other changes. Moreover, by helping to translate mRNA transcripts that are already present in cells, it enables proteins to be produced rapidly in response to stress, bypassing the need for a fresh round of transcription.
Last year, the same group revealed the importance of U1 (a major spliceosome component) in preventing premature termination of mRNA transcription. U1 deficit shortens mRNAs by proximal polyadenylation in 3′ UTR and introns. The mode of action is now known as “telescripting”.

■ Younis et al (2013) Minor introns are embedded molecular switches regulated by highly unstable U6atac snRNA. eLife, 2:e00780
■ Berg et al (2012) U1 snRNP Determines mRNA Length and Regulates Isoform Expression. Cell, 150: 53—64
Editorial coverage : Merkhofer et al (2012) U1 snRNA Rewrites the “Script”. Cell, 150: 9—11
cesarappleseed liked this
biochemistries posted this